
The Reactome Pathways database
Reactome is an open-source, curated, and peer-reviewed online pathway database for reactions in biological processes. The Reactome project aims to understand bioreaction systematically in humans collaborated by groups at the Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University School of Medicine, and The European Bioinformatics Institute.
Since the Reactome database is curate based on humans, other 19 non-human species in the last release of the Reactome database (Version 76 21 March 2021) are generating using orthology-based inferences methods. Reactome pathways are composed of different reactions, and the basic unit of reaction in the Reactome database is protein, metabolite, and complex (contains several proteins and metabolites) (Figure 1).
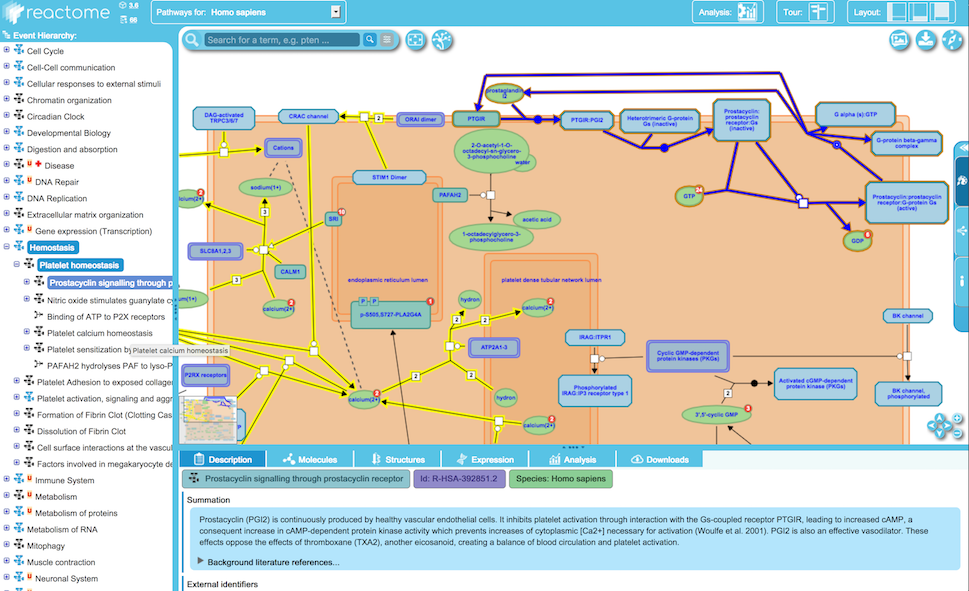
Figure 1: Example for a Reactome subpathway.
Each pathway in Reactome is identified by a stable ID that starts with R (Reactome), and the specie abbreviation (e.g., homo Sapiens: HSA) ends with a unique ID.
The Reactome pathways layout comprises six elements: compartments, nodes, edges, links, notes, and shadows(Figure 2).
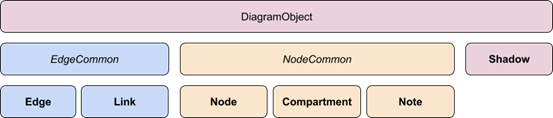
Figure 2: Graphic explanation for hierarchy relationship of the Reactome layout elements.
To better understand the functionality of those elements, please go and visit: https://reactome.org/dev/diagram/pathway-diagram-specs