
Metabolite class activity analysis
Metabolomics experiments might be targeted to one or several metabolite classes for which an assumption of change exists. For example, in experiments evaluating growth, the hypothesis of central carbon metabolites changing their levels may lead to the measurement of this type of compound only. In this scenario, most of the measured entities might show a significant change, which impairs the application of traditional enrichment methods that rely on a non-change background. On the other hand, pathway enrichment for metabolomics data rarely results in significant results as few pathway compounds are frequently successfully measured.
To test the hypothesis of a metabolite class being regulated, PaintOmics 4 implements a Metabolite Class Activity analysis tool, where a binomial test is used to assess the hypothesis of the proportion of significant compounds in a given measured metabolite class being higher than a user-defined threshold (Figure 1). If the user does not define an activity threshold, PaintOmics will use the average percentage of significant metabolites as the threshold for the "Generate automatically". P-values are corrected for multiple testing. These novel metabolomics analysis tools are provided as a separate tab in the main PaintOmics results panel. These visualisation tools enable easy navigation between metabolite data, neighbouring genes and metabolic pathways.
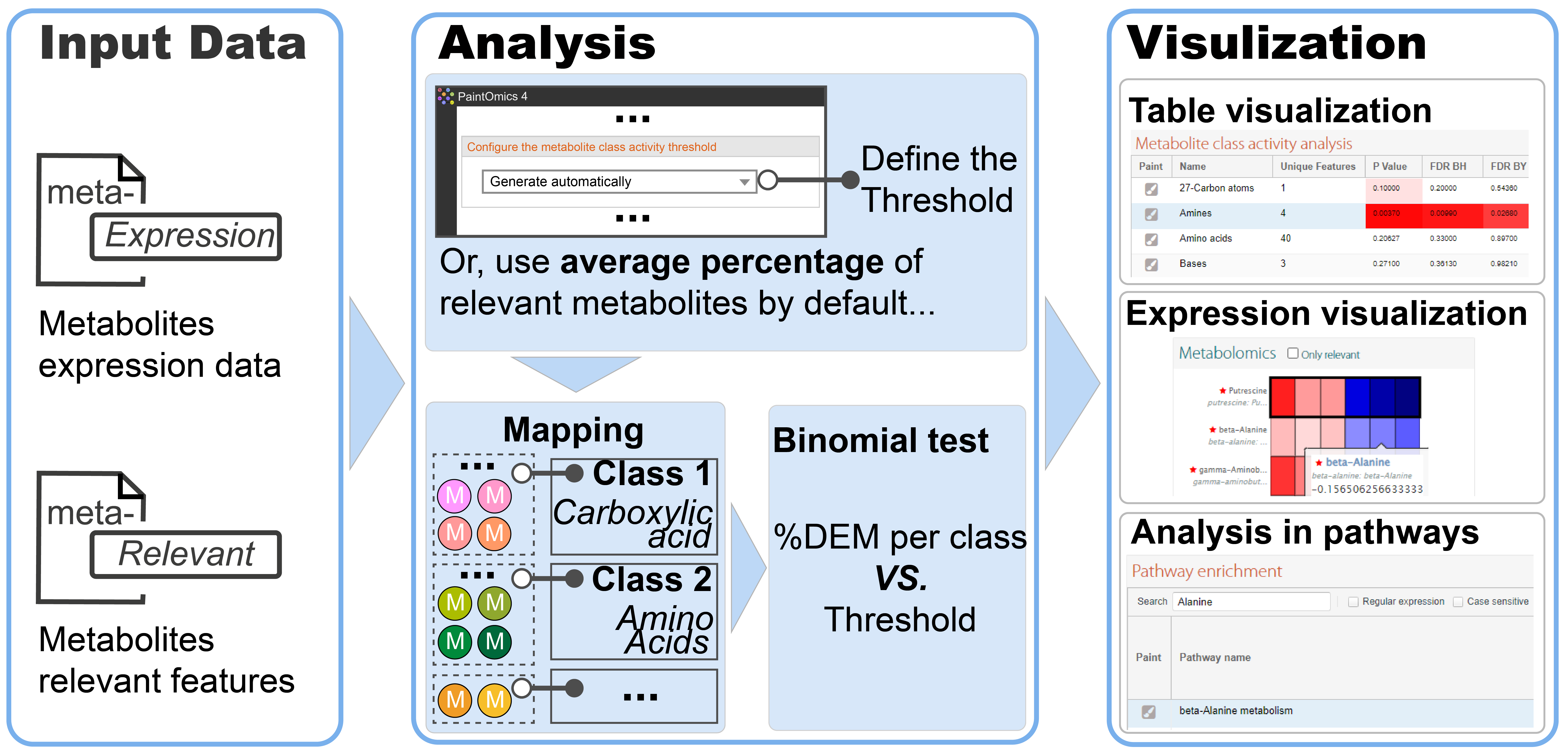
Figure 1: metabolite class activity analysis pipline in PaintOmics 4. DEM: Differentially Expressed Metabolite.