
The MapMan Pathways database
MapMan is a tool for multi-omics data annotations in the plant sciences based on plant-specific MapMan ontology. The ontology is organised in a hierarchical tree of biological concepts, which describe gene functions. MapMan group updated the previous MapMan to MapMan webserver biological pathway maps database named GoMapMan (http://www.gomapman.org/) (Figure 1).
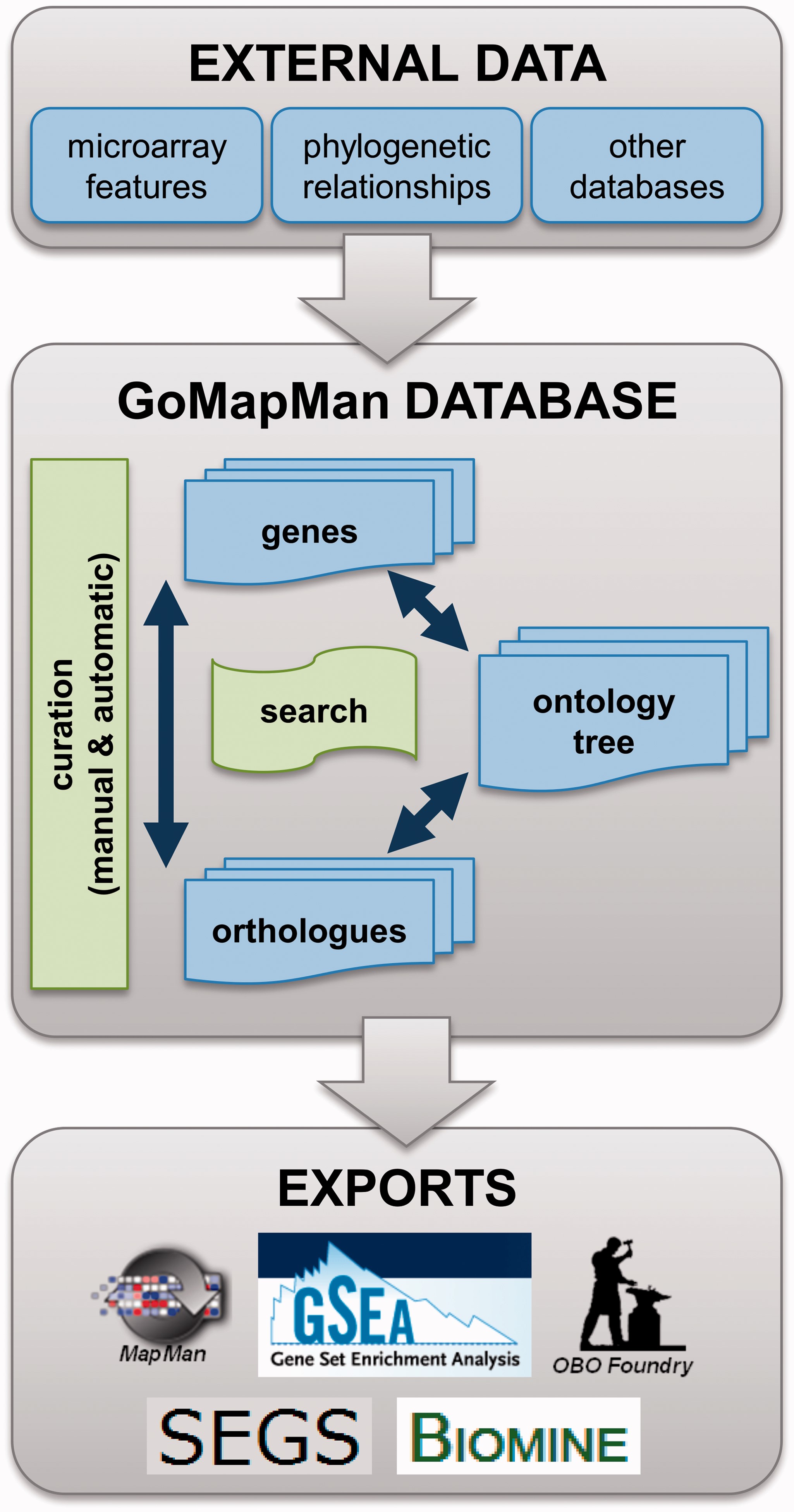
Figure 1: Schematic representation of GoMapMan database functionalities. The data are retrieved from various external sources. Genes are placed into the MapMan ontology tree and connected within a species or between different plant species via orthologue groups. Database contents can be searched or modified by manual or batch curation procedures and are furthermore available in the form of export files for different high-throughput analysis systems.
In GoMapMan biological pathway maps, there are three subsets:
- Protein GoMapMan: Protein-coding genes of various plants described with MapMan ontology terms, grouped according to various implemented orthologue groups.
- Metabolite GoMapMan: Plant metabolites functionally annotated with MapMan ontology terms.
- SmallRNA GoMapMan: Plant small RNA's functionally annotated with MapMan ontology terms. Currently only available for potato.